index.md 11KB
title: DNA Chisel - a versatile sequence optimizer
url: https://edinburgh-genome-foundry.github.io/DnaChisel/
hash_url: e8ebcfcc0d
DNA Chisel (complete documentation here) is a Python library for optimizing DNA sequences with respect to a set of constraints and optimization objectives. It comes with over 15 classes of sequence specifications which can be composed to, for instance, codon-optimize genes, meet the constraints of a commercial DNA provider, avoid homologies between sequences, tune GC content, or all of this at once!
DNA Chisel also allows users to define their own specifications in Python, making the library suitable for a large range of automated sequence design applications, and complex custom design projects. It can be used as a Python library, a command-line interface, or a web application.
Example of use
Defining a problem via scripts
In this basic example we generate a random sequence and optimize it so that
It will be rid of BsaI sites.
GC content will be between 30% and 70% on every 50bp window.
The reading frame at position 500-1400 will be codon-optimized for E. coli.
Here is the code to achieve that:
from dnachisel import * # DEFINE THE OPTIMIZATION PROBLEM problem = DnaOptimizationProblem( sequence=random_dna_sequence(10000), constraints=[ AvoidPattern("BsaI_site"), EnforceGCContent(mini=0.3, maxi=0.7, window=50), EnforceTranslation(location=(500, 1400)) ], objectives=[CodonOptimize(species='e_coli', location=(500, 1400))] ) # SOLVE THE CONSTRAINTS, OPTIMIZE WITH RESPECT TO THE OBJECTIVE problem.resolve_constraints() problem.optimize() # PRINT SUMMARIES TO CHECK THAT CONSTRAINTS PASS print(problem.constraints_text_summary()) print(problem.objectives_text_summary())
DnaChisel implements advanced constraints such as the preservation of coding
sequences, or the inclusion or exclusion of advanced patterns (see
this page
for an overview of available specifications), but it is also easy to implement
our own constraints and objectives as subclasses of dnachisel.Specification.
Defining a problem via Genbank features
You can also define a problem by annotating directly a Genbank as follows:

I this record:
Constraints (colored in blue in the illustration) are features of type
misc_featurewith a prefix@followed by the name of the constraints and its parameters, which are the same as in python scripts.Optimization objectives (colored in yellow in the illustration) are features of type
misc_featurewith a prefix~followed by the name of the constraints and its parameters.
The file can be directly fed to the web app or processed via the command line interface:
# Output the result to "optimized_record.gb"
dnachisel annotated_record.gb optimized_record.gb
Or via a Python script:
from dnachisel import DnaOptimizationProblem problem = DnaOptimizationProblem.from_record("my_record.gb") problem.optimize_with_report(target="report.zip")
By default, only the built-in specifications of DnaChisel can be used in the annotations, however it is easy to add your own specifications to the Genbank parser, and build applications supporting custom specifications on top of DnaChisel.
Reports
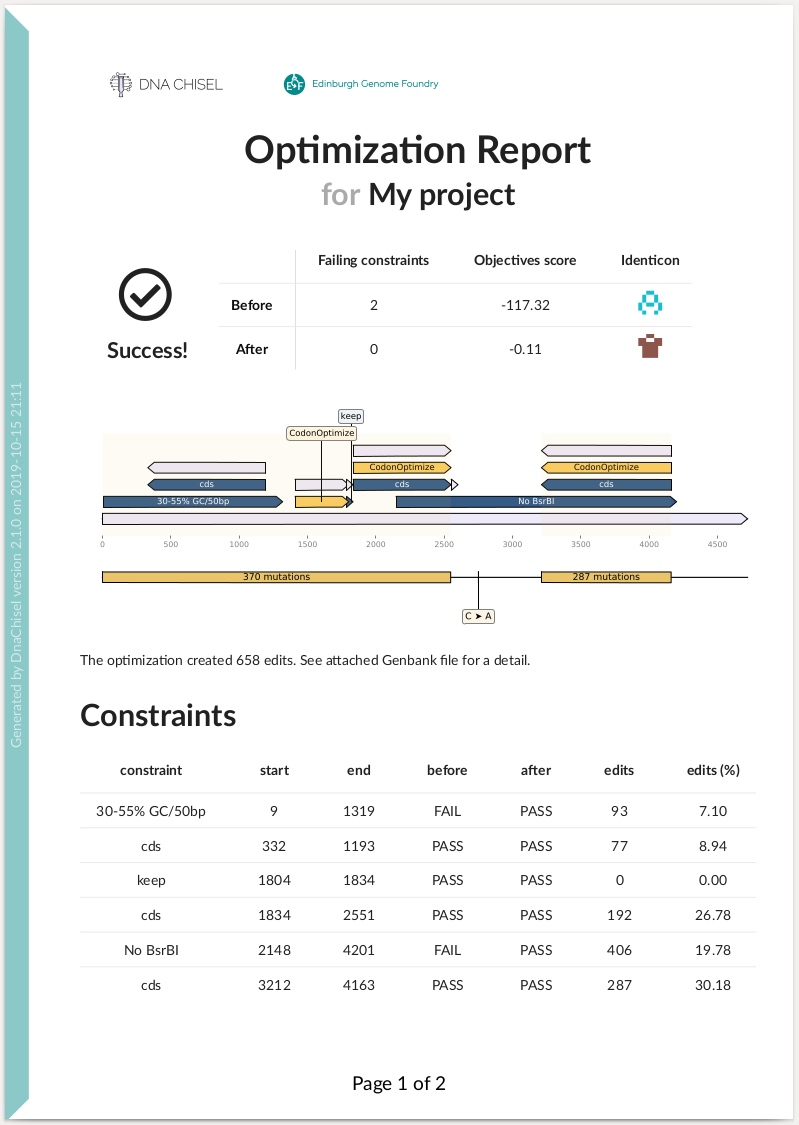
DnaChisel also implements features for verification and troubleshooting. For instance by generating optimization reports:
problem.optimize_with_report(target="report.zip")
Here is an example of summary report:
How it works
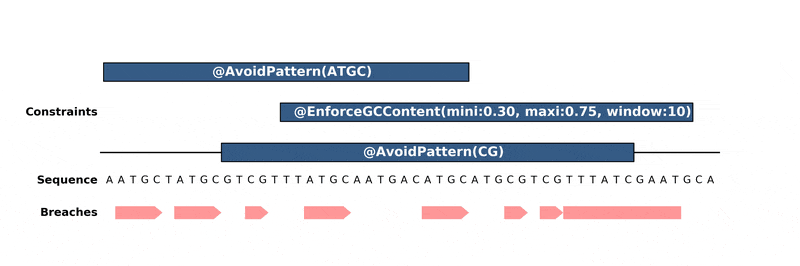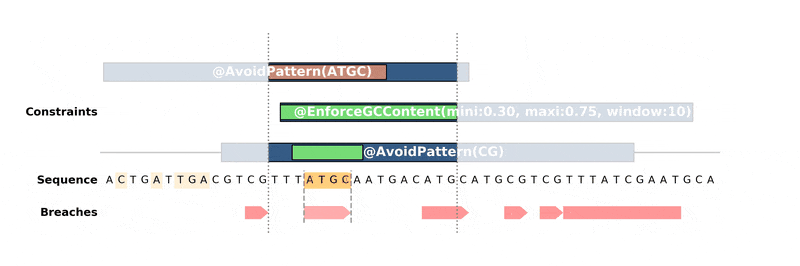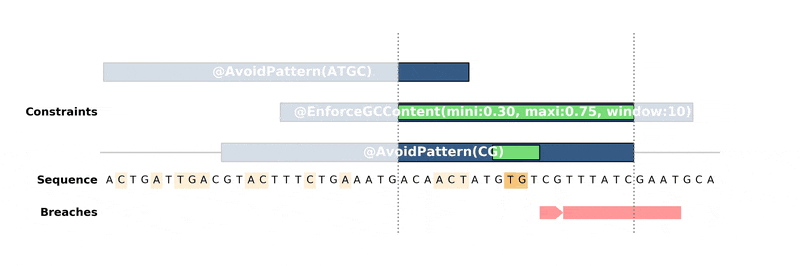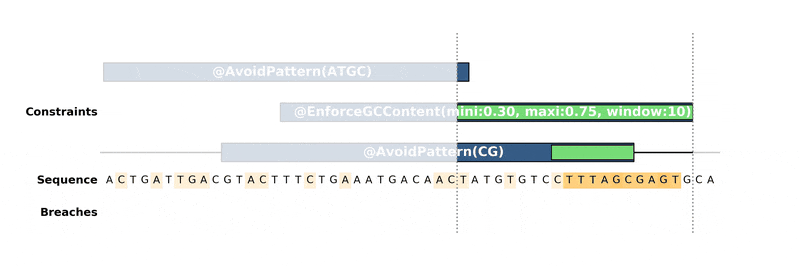
DnaChisel hunts down every constraint breach and suboptimal region by recreating local version of the problem around these regions. Each type of constraint can be locally reduced and solved in its own way, to ensure fast and reliable resolution.
Below is an animation of the algorithm in action:
Installation
You can install DnaChisel through PIP:
sudo pip install dnachisel[reports]
The [reports] suffix will install some heavier libraries
(Matplotlib, PDF reports, sequenticon) for report generation,
you can omit it if you just want to use DNA chisel to edit sequences and
generate genbanks (for any interactive use, reports are highly recommended).
Alternatively, you can unzip the sources in a folder and type
sudo python setup.py install
Optionally, also install Bowtie to be able to use AvoidMatches (which
removes short homologies with existing genomes). On Ubuntu:
sudo apt-get install bowtie
More biology software

DNA Chisel is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.
